Even C&EN reported the recent standardisation efforts by the oligosaccharide community on symbols to use for glycan* depiction. These guidelines are available online in Appendix 1B of Essentials of Glycobiology and will be updated over time.
As a test case for Sugar & Splice support, I depicted the oligosaccharide below whose structure is strangely reminiscent of Table 1 in the guidelines. For those of you glycan enthusiasts who wish to print T-shirts with this emblazoned on the front, here is an Inkscape-friendly SVG file.

However, such a diverse set of monosaccharide symbols is not present in the typical oligosaccharide. I’ve searched PubChem for the entry with the most symbols and found CID91850542 below with 11. (For an alternative depiction of the same structure, see GlyTouCan. Interestingly, the CSDB entry for the same paper describes a different but very similar glycan.):

In fact, having many symbols often indicates a dodgy structure as in the following example (PubChem CID101754793) deposited by Nikkaji which has 9 monosaccharide symbols. Looking at the original source, the SMILES not only has nitro as [N+](=O)O (must have been corrected by PubChem) but many of the sugars have incorrect stereochemistry (compared to the provided IUPAC name). The D/L in several of the symbols, indicating the presence of the rarer stereoisomer, is also a red flag.
 If you put the IUPAC name through OPSIN (after a minor mod), and then depict the resulting SMILES using Sugar & Splice, you get the correct structure:
If you put the IUPAC name through OPSIN (after a minor mod), and then depict the resulting SMILES using Sugar & Splice, you get the correct structure: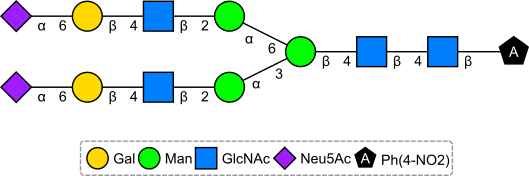
* Glycans are “compounds consisting of a large number of monosaccharides linked glycosidically”, via Wikipedia and the IUPAC gold book.